Abstract

Background: Systemic mastocytosis (SM) is a hematologic neoplasm characterized by the infiltration of clonal mast cells in the bone marrow or other extra-cutaneous organs. The clinical course varies between advanced and non-advanced (indolent and smoldering SM) forms of SM. The vast majority of patients harbor the activating D816V mutation in the KIT tyrosine kinase. Additional somatic mutations in other genes have been recognized as risk factors in SM. Cytogenetic aberrations are rarely found in SM but have been associated with advanced disease. Whole genome sequencing (WGS) and whole transcriptome sequencing (WTS) have been described as an alternative to cytogenetics and targeted molecular genetic analysis in myeloid cancers.
Aim: To assess the ability of WGS/WTS to detect cytogenetic aberrations and recurrent somatic mutations in SM.
Methods: 120 patients (51 female, 69 male) diagnosed with SM were analyzed with WGS/WTS and results were compared with orthogonal data of KIT D816V PCR, targeted sequencing, and cytogenetics. 47 patients (39%) were diagnosed with advanced SM (1 mast cell leukemia, 3 aggressive SM, 43 SM with associated hematologic neoplasm). For WGS, 2x151bp paired-end reads were generated on NovaSeq 6000 and HiSeqX machines (Illumina, San Diego, CA). BaseSpace's Tumor/Normal app v3 was used to call variants with Strelka Somatic Variant Caller v2.4.7 and structural variants (aberrations with >50bp in size) with Manta (v0.28.0). Genomic DNA from a mixture of multiple anonymous donors (Promega, Fitchburg, WI, USA) was used as normal. For WTS, 2x101 bp paired-end reads were produced with a median of 50 mio. reads per sample, aligned with STAR v2.5.0, and variants were called using Isaac Variant Caller v2.3.13.
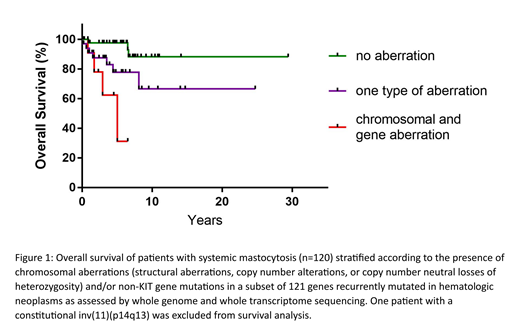
Results: WGS/WTS detected cytogenetic aberrations in 21% of patients: 2 patients displayed a complex aberrant karyotype, 3 balanced structural aberrations, 16 copy number alterations, and 6 copy number neutral losses of heterozygosity. Aberrations detected by chromosome banding analysis were also found by WGS in all but three patients (small clones with aberrations present in ≤20% of metaphases and <10% of interphase nuclei as determined by FISH). In contrast, WGS/WTS detected additional aberrations in 16 patients. The frequency of chromosomal aberrations detected by WGS/WTS was higher in advanced compared to non-advanced SM (34% vs. 12%, p<0.05). KIT D816V was detected by PCR in 98%, by WGS in 21% and by WTS in 46% of patients. The detection rate by WGS was significantly higher in advanced (36%) compared to non-advanced SM (12%, p<0.05) while no difference was observed for WTS (45% vs. 47%). Somatic mutations outside of KIT were analyzed within a subset of 121 genes recurrently mutated in hematologic neoplasms. 46% of patients showed non-KIT mutations with a median of 2 mutations per patient. Both frequency of non-KIT mutations as well as the median number of mutations per patient was higher in advanced (83%; n=3) compared to non-advanced SM (22%, n=1, p<0.05). Finally, we analyzed the impact of genetic aberrations on survival in our SM cohort. Patients were grouped according to the presence of chromosomal aberrations and gene mutations (non-KIT) as assessed by WGS/WTS. SM patients with both types of aberrations (n=16), one type of aberration (n=47; gene mutations only n=38; chromosomal aberrations only n=8), or no aberration but KIT D816V (n=57) showed significant differences in overall survival (p<0.05, Figure 1).
Conclusions: WGS/WTS has limited sensitivity for detection of KIT D816V in SM. This finding can be explained by the low KIT D816V mutation burden typically found in bone marrow aspirates of SM patients. In line, we observed a slightly higher detection rate in advanced SM and in RNA-based WTS analysis. As WGS/WTS will be applied for the diagnostic workup of myeloid malignancies in the future and SM associated with other hematologic neoplasms may be overlooked if not specifically investigated, additional PCR-based techniques are still mandatory to rule out KIT D816V as a diagnostic criterion for SM. In contrast, WGS/WTS detects both chromosomal aberrations and additional gene mutations in patients with SM and can be used as an alternative to cytogenetics and targeted sequencing for risk assessment. In particular, the absence of genetic aberrations in WGS/WTS identifies SM patients with indolent course of the disease and favorable prognosis.
Hoermann: Novartis: Honoraria. Kern: MLL Munich Leukemia Laboratory: Other: Part ownership. Haferlach: MLL Munich Leukemia Laboratory: Other: Part ownership. Haferlach: MLL Munich Leukemia Laboratory: Other: Part ownership.
Author notes
 This icon denotes a clinically relevant abstract
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal